The analysis in this tutorial is typical of experiments in eukaryotic species with high-quality genomes and genome annotation available. Currently Ilumina sequencing produces longer reads for which the new version of Tophat should be used version 130.
Bioinformatics Greifswald Incorporatingrnaseq Tophat
Press the esc key to exit insert mode.

. Tophat is a splicing aware aligner so we can map transcripts to the genome. Tophat is a splice-aware mapper for RNA-seq reads that is based on Bowtie. Critically the number of short reads generated for a particular RNA is assumed to be proportional to the.
In this tutorial well map reads from an RNA-seq study in Drosophila melanogaster to the reference genome using tophat. To install TopHat from source package unpack the tarball and change directory to the package directory as follows. This is a practical hands-on tutorial designed to give participants experience with RNA-Seq data analysis using Tophat Cufflinks and CummRbund in Galaxy.
A FASTQ file normally uses four lines per sequence. Getting started The main goal of this activity is to go through a standard method to obtain gene expression values and perform differential gene expression analysis from an RNA. Here we describe the method of analyzing RNA-seq data using the set of open source software programs of the Tuxedo suite.
TopHat and Cufflinks provide a complete RNA-seq workflow but there are other RNA-seq analysis packages that may be used instead of. Jeremy Goecks Galaxy RNAseq tutorial httpmaing2bxpsueduujeremypgalaxy-rna-seq-analysis-exercise. Tophat -o output_directory -p1 genome_database rnaseqfastq.
The newest version of Tophat can be invoked just as tophat in the command line without version string. To understand the basics of RNA-Seq data how to use RNA-Seq for different objectives and to familiarize yourself with some standard software packages for such analysis. Go to an empty line with you cursor and copy paste the new RNA_HOME and PATH commands into the file.
Line 2 is the raw sequence letters. Line 1 begins with a character and is followed by a sequence identifier and an optional description like a FASTA title line. Line 3 begins with a character and is optionally followed by the same sequence identifier and any description again.
A set of lectures in the Deep Sequencing Data Processing and Analysis module will cover the basic steps and popular pipelines to analyze RNA-seq and ChIP-seq data going from the raw data to gene lists to figures. Transcriptome splice-variantTSSUTR analysis microRNA-Seq etc. It uses the mapping results from Bowtie to identify splice junctions between exons.
This FASTQ files are RNA-seq data from two samples. Map the reads to reference genome using TOPHAT. You first need to build an index file for your genome.
Filtering raw alignments step 1 Tophat produced an bam file at output_directoryaccepted_hitsbam. We recommend that you watch the video Aligning RNA-seq reads to reference genome instead which covers t. There are several types of RNA-Seq.
This tutorial from 2017 covers the TopHat aligner. The requirements for aligning this type of data is slightly different from eg. Background Web Resources.
Press the key to enter command mode. This tutorial will focus on doing a 2 condition 1 replicate transcriptome analysis in mouse. RNA-seq transcriptome sequencing is a very powerful method for transcriptomic studies that enables quantification of transcript levels as well as discovery of novel transcripts and transcript isoforms.
More information on Tophat can be found here. TOPHAT is widely used in the early days of RNA-seq data analysis. Some of the applications used in RNA sequencing analysis are the following.
It is slow but consumes less memory. If you would like to learn more about how to use vi try this tutorialgame. Much of Galaxy-related features described in this section have been developed by.
Align the RNA-seq short reads to a reference genome. MRNA rRNA miRNA converting in some way to DNA and sequencing on a massively parallel sequencing technology such as Illumina Hiseq. Once installed run the plugin by selecting your reads and reference sequence then clicking on AlignAssemble - Map to Reference in the toolbar.
The real RNA-seq data would normally take several hours to process. STAR is much faster but need a machine with large memory 30GB for human genome. TopHat2 uses using Bowtie to align RNA-Seq reads to mammalian-sized genomes and then analyzes the mapping results to identify splice junctions between exons.
TopHat is designed to align RNA-seq reads to a reference genome while Cufflinks assembles these mapped reads into possible transcripts and then generates a final transcriptome assembly. Configure the package specifying the install path and the library dependencies as needed. These lectures also cover UNIXLinux commands and some programming elements of R a popular freely available statistical software.
Afterwards align the RNAseq data to the genome. RNA-seq as a genomics application is essentially the process of collecting RNA of any type. Press the i key to enter insert mode.
If you are not in working directory already type cd workdirusre_user_ID first ls. This is quite different conceptually to mapping to the transcriptome directly. Bowtie1 is an ultrafast memory-effi cient aligner for large sets of short reads.
Previous version of Tophat 120. Type wq to save and quit vi. This tutorial is inspired by an exceptional RNA seq course at the Weill Cornell Medical College compiled by Friederike Dündar Luce Skrabanek and Paul Zumbo and by tutorials produced by Björn Grüning bgruening for Freiburg Galaxy instance.
This practical will introduce some popular tools for basic processing of RNA-seq data. At the very end we can compare these results to the results we got from mapping directly to the. RNA-Seq Tutorials Tutorial 1 RNA-Seq experiment design and analysis Instruction on individual software will be provided in other tutorials Tutorial 2 Hands-on using TopHat and Cufflinks in Galaxy Tutorial 3 Advanced RNA-Seq Analysis topics.
Inspect the files in the working directory workdirmy_user_ID. Special files were prepared for this workshop so that the exercise can be finished in minutes The files only include reads from first 20mb region from a genome. Quick Start Install the plugin by downloading the gplugin file and dragging it in to Geneious or use the plugin manager in Geneious under Tools - Plugins in the menu.
The Cufflinks Rna Seq Workflow
Basic Analyses With Tophat Cufflinks Rnaseq Tutorial 1 Documentation
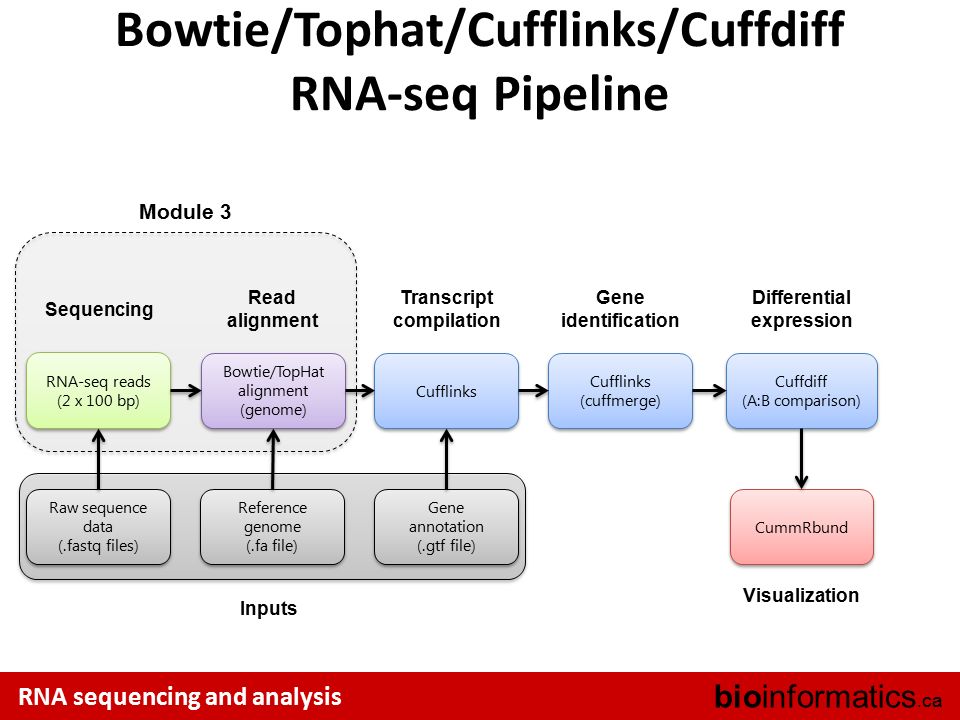
Canadian Bioinformatics Workshops Ppt Video Online Download

Rna Seq Course Alignment Using Tophat Old Youtube
Aligning Rna Seq Data Ngs Analysis
Tophat Cufflinks Command Pipeline

Reference Based Rnaseq Data Analysis Long
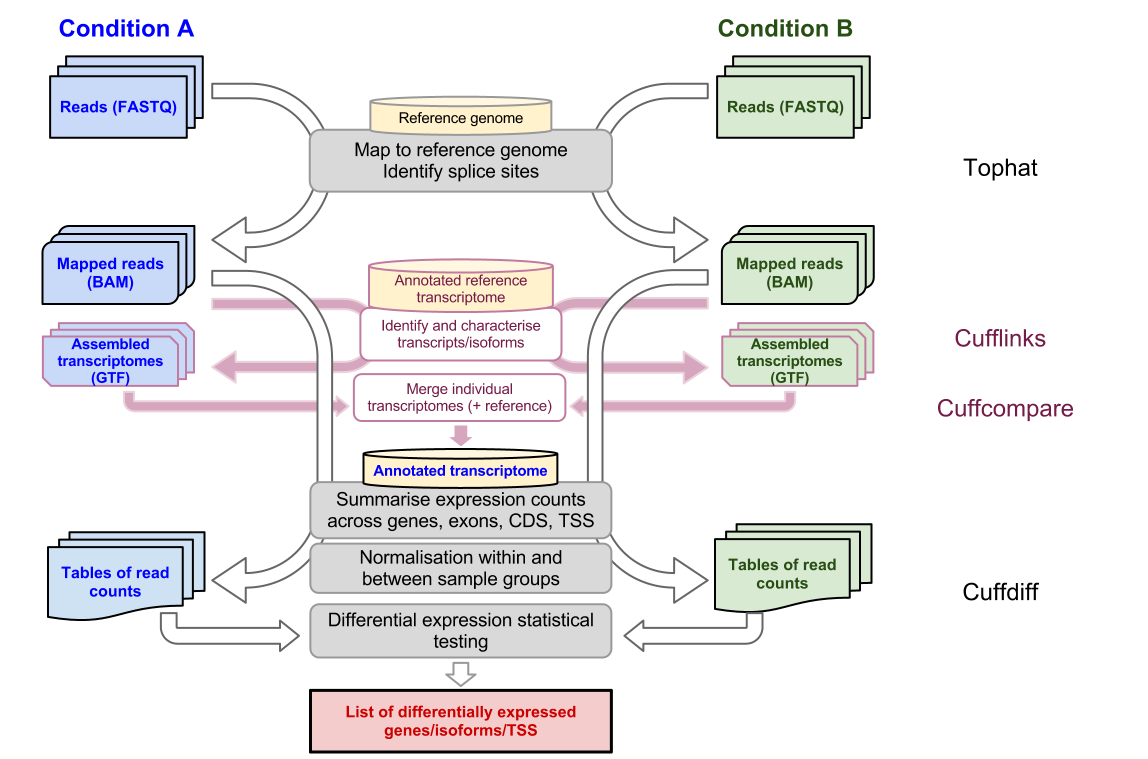
Introduction To Bulk Rnaseq Analysis Bioinformatics Documentation
0 comments
Post a Comment